
For instance to get the complete overview of the biologic’s stability, function, and impurity profile. Send us a request if you would like us to prepare a custom project package. to fully characterize the biopharmaceutical product. We often combine protein sequencing with intact mass analysis, host cell protein analysis, etc. But we can also make peptide maps without protein sequence information. For quantitative peptide maps we mainly use MS technology to search specific protein databases. The Edman degradation analysis provides up to 40 amino acid residues from the N-terminal – without any prior knowledge about the protein sequence. Should you use Edman protein sequencing or mass spectrometry? Therefore, the protein sequencing service provided by Alphalyse is based on the original Edman degradtion method – and high-resolution mass spectrometry technology.
Protein sequence analysis full#
But sequencing approaches must be robust, reproducible and sensitive to obtain full sequence coverage. The primary structure of a protein determines how the protein folds into a unique three-dimensional structure secondary, tertiary, and quaternary structures, which in turn determines the biological function of the protein. Initiation and termination of the ion/ion reaction is controlled by adjustment of these dc bias potentials.Do you want to identify or confirm the amino acid (AA) sequence of your protein, antibody, or peptide? Do you need a service to pinpoint truncations or cleavage sites? Or even quantify post translational modifications?Īmino acid sequencing of proteins, antibodies, and peptides play an important part of detailed sequence analysis and protein characterization. Protein Sequence Analysis Service Each protein has a characteristic number and sequence of amino acid residues. 4, manipulation of the dc bias potentials, applied to the QLT's segments and end lenses, permits axial segregation of precursor cations and reagent anions during anion injection and isolation.
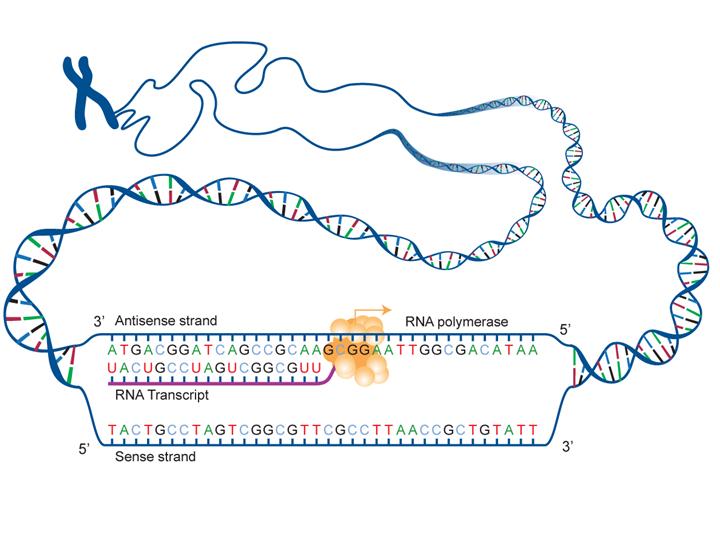
HISTORY 1951: The very first sequence of insulin protein were characterized by Fred Sanger. Imp in targeting drugs to specific metabolic pathways. Imp for understanding cellular functions. The QLT instrument has several unique advantages over QIT instruments for performing ion/ion experiments, including greater ion capacity (≈30-fold) and higher ion-injection efficiency (≈10- to 30-fold) ( 39). Protein Sequencing: Technique to find out amino acid sequences in protein. Brutto formula and length of the protein are calculated always (independently of items you have chosen). This conclusion is supported by the finding that m/ z 179 is not observed when argon is used as the CI reagent gas (data not shown). Peptide Protein Sequence Analysis: Purpose: Copy or type amino acid sequence of a protein, choose necessary items in 'options' and push button 'Calculate'. The target sequence databases we currently offer include UniProtKB/TrEMBL (uniprottrembl), UniProtKB/Swiss-Prot (uniprotsprot), the protein sequences from the PDB (pdbnr), and the nonredundant protein sequence database (nr).
We assume that the latter species is formed by a two-step process that involves electron capture and hydrogen atom abstraction from methane (Eqs. Select a target protein sequence database in the drop-down list over which you wish to build the alignment (Fig. On top of our advanced technologies in bioinformatics, we combine protein. 1.The main feature of email protected is the interconnection of all methods gathered within a simple and user-friendly Web interface. One pathway for this process involves generation of an odd-electron hypervalent species ( \(\begin\), respectively. General protein sequence databases, sequence similarity search and alignment tools (77) Individual protein families (78) Protein domains, classification and. Creative BioMart, with a successful track record of offering more than ten thousand custom bioinformatics consultations, provides protein sequence analysis of proteins by classifying them into families and predicting domains and important sites. The general flowchart describing relationships between biological sequence data and biocomputing methods available on the email protected web server is given in Fig. Capture of a thermal electron by a protonated peptide is exothermic by ≈6 eV (1 eV = 1.602 × 10 -19 J) and causes the peptide backbone to fragment by a nonergodic process, e.g., one that does not involve intramolecular vibrational energy redistribution ( 2– 5). In this method, multiply protonated peptides or proteins are confined in the Penning trap of a Fourier transform ion cyclotron resonance (FTICR) mass spectrometer and exposed to electrons with near-thermal energies. Six years ago, McLafferty and coworkers ( 1) introduced a unique method for peptide/protein ion fragmentation: electron capture dissociation (ECD).


 0 kommentar(er)
0 kommentar(er)
